
Written by the NTNU iGEM team together with Rahmi Lale.
The Giant Jamboree competition 2014 is only two days away. Hynes Convention Center in Boston is hosting the event on 30 October – 3 November. Here, 2.500 Synthetic Biology researchers from 245 universities in 32 countries are participating. One of these teams is from NTNU.
An iGEM team from the NT faculty
The 2014 iGEM team from NTNU consists of eight students and three instructors. This year all students are part of the NT faculty, and everyone is either a student of the five-year master or three-year PhD program in biotechnology. Four of us (Camilla M. Reehorst, Line Aa. Omtvedt, HyeWon Lee and Ronja Hesthammer) finished our master program this year, and we have experienced the work with iGEM exciting and educational. Having a master’s degree does not mean you are finished learning, and we think that for most of us the “soup bowl of knowledge” has only just begun to be filled. These four students all undertook the laboratory part of their master’s at the Department of Biotechnology under the biopolymer chemistry branch. Not all of us have attained a job yet, but with the iGEM experience under our wings, the prospects seem a tad brighter. One of us (Eivind B. Drejer) is a fourth year student. He started to swim with the rest of the master student submarines, fully submerged in literature, in August 2014. His master thesis is under the systems biology branch of biotechnology (meaning a lot of computer fiddling), and he has not scheduled laboratory work in his thesis. His iGEM experience is therefore considered to supplement his degree to the point where future employers will tear at each other’s throats for such diverse experience in their workers.
Two of us (Pål Røynestad and Elias H. Augestad) are third year students, and have not yet started work on master’s thesis. Pål is intrigued by the systems biology branch, and will start a computational and modelling-based assignment this autumn. Elias, on the other hand, is interested in molecular biology, and will start a master’s degree at the University of Copenhagen in September 2014. They might be the least experienced (based purely on age and amount of years spent studying biotechnology), but they are very valuable assets to our team. We would be lacking without them. Our last team member (Jacob Lamb) is a PhD student, and out-ranks us all. His field is molecular biology, and he has worked extensively with photosynthetic organisms. Not only does he actually know what he is doing in the lab, he is also a native English speaking type of guy, which means it sounds like he knows what he’s doing (unlike the rest of us that express ourselves with communication stutter steps). We also have three amazing instructors – Eivind Almaas, Rahmi Lale and Martin Hohmann-Mariott. They have participated in this competition before, both as instructors and judges. Their experience and knowledge is sorely appreciated, and to top it off they are very helpful!

Creating a puzzle from BioBricks
Every year the iGEM foundation sends each participating team packets brimming with what are known as BioBricks. These are biological parts that teams can use in their respective projects. The BioBricks are DNA sequences with specific functions that can be assembled to form a variety of different constructs. The amount of BioBricks in the iGEM registry usually increases from year to year, since each participating team has the opportunity to send in and register new and exciting parts. The BioBricks arrive in small metallic packages that look unmeritedly plain.
Inside, however, is where the magic lies. As illustrated by Elias, opening and examining the ordinary metal plate contents leaves a person smiling and happy.

The DNA sequences in each well are dried before they are shipped, and in order to use them, the DNA must be resuspended in distilled water. Once this is done, the DNA can be transferred into a culture of competent bacteria where the BioBrick parts multiply along with their hosts. Sometimes the BioBricks contain markers, meaning that the transformation success rate can easily be measured. Instead of laborious plating of negative non-transformed bacteria, the researcher can simply look for red cultures (given that the marker was a fluorescent gene that turns the bacteria red) as seen in the picture below.
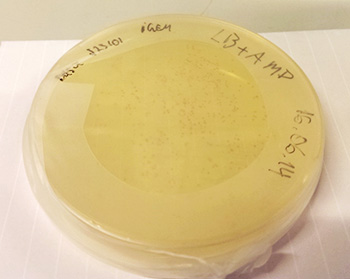
In our project, we wish to use some of these BioBricks to create an inducible expression system in Synechocystis sp. PCC 6803. Doing this is like building a puzzle; every piece belongs to a specific location, and if one piece is missing the whole image is obscured. Adding one BioBrick to another is not necessarily a time consuming process, but because the puzzle consists of many pieces we will need to spend some time getting the final construct together. The process involves enzymatic PCR amplification and ligation (Gibson assembly method), with antibiotic-based selection on agar plates. We recently managed to create our desired construct in E. coli DH5α cells.
The visible marker is a red fluorescent protein, and as can be seen in the picture below, the colonies casts a red hue. When the inducible expression system is functional, we wish to use this to up-regulate genes that might cause the bacteria to fixate more carbon, with the overall aim of the project is to decrease carbon dioxide emissions from factory exhaust.
We are planning on up-regulating glucose oxidase in Synechocystis in order to achieve this. Glucose oxidase is an enzyme that exhausts the oxygen level in our bacteria. The idea is that the bacteria will start metabolising more carbon dioxide once the oxygen is depleted; however, the growth rate of the bacteria will most likely suffer with our intervention, and we therefore hope that the carbon dioxide uptake is greater than the loss in growth rate compared to a wild-type Synechocystis culture. We are excited to see how well our construct will develop, and will rejoice the day our instrument is implemented in factory chimneys.
The realistic hope is that we receive a gold medal at the iGEM world final in Boston, Massachusetts in October-November 2014. Gold medals are given to any team that satisfy a set of criteria and quality standards determined by the iGEM administration. These criteria can be tricky, but we are confident in our abilities to achieve our goals. In addition to bronze, silver and gold medals, teams compete for special awards for a variety of contributions. These awards are only given to one team each year, and are therefore a symbol of greatness. The NTNU_Trondheim iGEM team 2014 has high ambitions and we aspire for greatness, but being humble we do not expect any special prizes. That being said, we would all like to proudly return home with a solid glass trophy in our greedy little hands.
